Koala Endogenous Retroviruses (ERVs) Protect against Retroviral Infections
I don’t really have a bucket list, but if I did, hanging out with koalas would most certainly be near the top.
My wife Amy and I had a chance to live out this “bucket list” experience a few years ago when we visited Australia. I must confess, it was a bit of a disappointment. Sure, the koalas were unbelievably adorable, but they smelled like cough drops and slept the entire time we were in the exhibit area.
Koalas smell like cough drops and sleep up to 20 hours a day because of their diet. These marsupials primarily consume eucalyptus leaves. This behavior is remarkable because the leaves are loaded with toxins and have little nutritional value. Some biologists think that koalas have a “super liver” that can detoxify the harmful components of the eucalyptus leaves. Others speculate that their gut microbiome metabolizes the toxins in their diet. Koalas spend most of their “waking hours” sleeping because it helps them conserve the little energy they glean from their diet. Some biologists believe that they also sleep for long periods of time because some of the compounds found in eucalyptus leaves are narcotics.
Retroviruses Threaten Koala Survival
Sadly, conservation biologists think that koalas are in danger of extinction. Habitat destruction along with brushfires and drought caused by climate change threaten these creatures. But so does an infectious agent, called the koala retrovirus (KoRV). Some biologists believe that this virus causes koala immunodeficiency syndrome (KIDS), which is similar to AIDS in humans. Infection with this virus leaves koalas unusually susceptible to pathogens and cancers. Biologists worry that the spread of this virus among koalas may be at the pandemic level.
Because of this concern, biologists have been urgently investigating the KoRV with the hope that the insight gleaned from studying this retrovirus can be used to stave off koala extinction. There is an added benefit to the work. They also hope that insights they gain by learning about the KoRV will yield a basic understanding of the biology of retroviruses, and specifically, the process by which retroviruses become endogenized. (I will elaborate on endogenization below.)
A Surprising Discovery with Surprising Implications
In 2018, a research team with contributors from the United States, Germany, and Australia reported on the discovery of a previously unknown defense mechanism that protects koala germ cells from retroviral infections.1 Remarkably, this defense mechanism relies on the endogenous retroviral sequences already present in the koala genome—sequence elements long thought to be junk DNA.
Not only does this discovery bear importance for conservation efforts, but it adds to the mounting evidence that—far from being a wasteland of nonfunctioning junk—genomes are highly sophisticated systems with most of the sequences serving critical functions.
This changing perspective on the architecture and operation of genomes satisfies a key prediction of the RTB genomics model, providing justification for creationists and intelligent design proponents who view genomes as the work of an intelligent agent.
I will elaborate on this point later. But first I’ll supply a bit of background on retroviruses and endogenous retroviruses.
How Retroviruses Infect Cells
Like all viruses, retroviruses consist of genetic material surrounded by a capsid. Retroviruses infect organisms by invading specific cell types of the host organism. After the retroviruses attach to the target cell’s surface, the targeted cell engulfs them. Once inside the targeted cell, the viral genetic material exploits its machinery to produce viral genetic material and viral proteins. Once formed, these biomolecules assemble into new viral particles. When these newly formed viruses escape from the invaded cell, the infection cycle repeats.
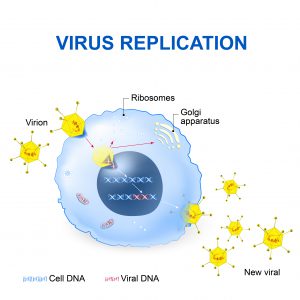
Title: Virus Replication
Credit: Shutterstock
For many retroviruses, their genetic material (RNA) is converted into DNA before the infectious cycle can proceed. This conversion is carried out by an enzyme called reverse transcriptase, delivered to the target cell along with the retroviral RNA. The enzyme uses the retroviral RNA to make DNA, which can then exploit the invaded cell’s biosynthetic operations to direct the production of new retroviral particles. The DNA copy of the retroviral genetic material can also become incorporated into the host cell’s genome, becoming part of the host cell’s genome. This process is called endogenization.
Endogenous Retroviruses (ERVs)
Once retroviral DNA becomes incorporated into an organism’s genome it is called an endogenous retrovirus (in contradistinction to exogenous retroviruses, which exist independent of genomes). After inserting into the host’s genome, the endogenous retrovirus can still produce retroviral particles, if its DNA is transcribed by the host cell’s biochemical machinery. If the ERV infects a germ line cell (a sperm cell or an egg cell), it can be inherited and, as a consequence, transmitted from generation to generation as a permanent feature of the genome.
If the ERV DNA undergoes severe mutations it becomes disabled, remaining in the genome as nonfunctional, junk DNA. As it turns out, about 8% of the human genome consists of what appears to be inactive ERVs.
ERVs as Mobile Elements in the Genome
ERVs possess an interesting capacity called retrotransposition. This capacity allows ERVs to make copies of themselves with the duplicate copies reinserting themselves in the genome. For this reason, life scientists refer to ERVs as mobile DNA elements. ERVs are also called transposons or retrotransposons. For ERVs, the process of retrotransposition begins when the ERV is transcribed by the host cell’s machinery into RNA. The enzyme reverse transcriptase converts the ERV RNA into DNA, which can be randomly inserted into the host cell’s genome through the activity of an enzyme called an integrase.
Endogenous Retroviruses and the Case for Human Evolution
Evolutionary biologists regard the endogenous retroviral populations that have become incorporated into the human genome as evidence that humans share an evolutionary history with the great apes. Many human ERVs are also found in the genomes of chimpanzees, bonobos, gorillas, and orangutans. Not only do these ERVs share many of the same sequence patterns, but they also appear in corresponding locations in the genomes.
Evolutionary biologists explain this data by assuming that the shared ancestor of humans and chimpanzees, for example, became infected by these specific retroviruses. Later these endogenized retroviruses underwent mutations that disabled them in the organism’s genome. These ERV sequences were then retained in the genomes of humans and chimpanzees as their separate evolutionary lineages diverged from the common ancestor. According to the model, the ERVs shared by humans and chimpanzees represent the molecular artifacts of infections that occurred millions of years ago and left their imprint on contemporary genomes via this (presumed) shared ancestor.
For many people, the presence of ERVs in the human genome (and the genomes of other organisms) is difficult to explain from a creation model perspective. In fact, I have interacted with several people who have told me that the distribution of ERVs in the genomes of humans and great apes is what convinced them that human evolution did, indeed, occur. After all, why would a Creator introduce the same nonfunctional sequence elements in the same locations within the genomes of organisms that naturally group together (based on other biological features)? And why would he create these shared sequence elements to bear such strong similarity to retroviruses?
On the other hand, the RTB creation model predicts that junk DNA sequences—including ERVs—are functional, serving a vital role. The model also predicts that a rationale exists for the sequence similarities between retroviruses and ERVs. Some recent insights on the KoRV provide a response to these two challenging questions.
The Koala Retrovirus
Biologists believe that ERVs in the human genome are ancient. Accordingly, the “youngest” human ERV sequences arose around 5 million years ago. For this reason, life scientists have expressed concern that they can’t “observe” and study the endogenization and inactivation processes that lead to ERVs. Because of this limitation, biologists have used the sequence features of ERVs and the behavior of retroviruses to infer the mechanistic steps that they believe led to the origin of ERV sequences in genomes.
In contradistinction, the exogenous KoRV appears to be actively invading koala genomes, generating the endogenous form of the retrovirus on an ongoing basis. Many of the endogenous KoRVs appear to be active. This scenario means that the KoRV is transmitted horizontally, from koala to koala, via the exogenous form of the retrovirus and vertically, from generation to generation, via the endogenous forms that have become incorporated into the germ cells.
Ancient Koala ERVs Protect the Koala Genome from KoRV
The research team from Germany, the US, and Australia detected variants of the endogenized KoRV in koala genomes that appear to be a combination of KoRV and other preexisting ERVs also found in the koala genome. In other words, it appears as if preexisting ERVs in the koala genome—through the process of retrotransposition—inserted copies of themselves into endogenized KoRV, disrupting their sequence and, consequently, disabling them. The researchers concluded that this process doesn’t appear to require the DNA sequence of the preexisting ERV to bear any similarity to the KoRV.
In other words, the research team discovered that preexisting ERVs play a role in imparting koalas with an innate immunity against KoRVs. In fact, the researchers believe that this mechanism may be widespread among animals, including humans. If so, the ERVs embedded within genomes serve as an antiviral defense system.
ERVs: Common Descent or Common Design?
Many life scientists regard the shared biological features possessed by organisms (that naturally cluster together) as evidence for their shared evolutionary ancestry. Yet, an alternative explanation can be posited for these biological similarities. Instead of evincing common descent, they could be interpreted as shared biological designs. In fact, prior to Charles Darwin, the prominent biologist Sir Richard Owen provided a theoretical framework to interpret anatomical and physiological similarities shared among organisms. Owen saw these mutual features as manifestations of a common blueprint—an archetype.
The RTB creation model picks up where Owen left off, viewing the shared features in the genomes of organisms as manifestations of genomic archetypes. In other words, the genetic similarities in the genomes of humans and the great apes were intentionally introduced by the Creator. To justify this interpretation, the shared genomic features must serve a function. The discovery that preexisting ERVs in the koala genome can disrupt the endogenization process of the KoRV—and the view that this process may be widespread among other organisms—satisfies this requirement.
Of course, this insight then prompts the question: Why would a Creator produce functional genomic features that so closely resemble an endogenized retrovirus? Again, insight from the work on the KoRV addresses this question. The structural and functional features of the preexisting ERVs (i.e., their capacity to copy themselves and move throughout genomes) are precisely what make these ERV sequences so useful. Their capacity for retrotranspositioning affords these sequences the means to disrupt the endogenization process of invading retroviruses. In other words, for the ERV sequences to operate as antiretroviral elements, they must resemble endogenized retroviruses.
If the creation model perspective on ERVs is valid, then it suggests that ERVs may protect the host cell’s genome from retroviral infections through other mechanisms, like competitive inhibition. Most ERV sequences, like retroviral genomes, consist of two noncoding regions on the 3´ and 5´ ends of the sequence called long terminal repeats (LTRs). The ERV sequences also contain genes for reverse transcriptase and the proteins located in the virus capsule. If the ERV sequence is transcribed to produce ERV RNA and if the capsid proteins are produced, then both the RNA and the capsid proteins could inhibit the assembly of invading retroviral particles, through competitive inhibition, which would prevent the transmission of the invading retrovirus to other cells. In this scenario, the similarity of the ERVs to retroviruses is crucial.
This proposed antiviral mechanism has been observed by researchers from Imperial College London in the UK.2 They detected a conserved ERV sequence in the genomes of humans, sheep, mice, and rats. They found this sequence to be under the influence of purifying selection. In other words, the sequence must be functional. It codes for proteins that are similar to those found in retroviruses. The researchers speculate that when these genes are transcribed and translated, they interfere with the assembly of retroviruses.
The bottom line: ERVs serve at least one function, accounting for their presence in genomes from a creation model vantage point. In light of this function, a rationale indeed exists for their structural and functional similarity to retroviruses.
Based on this similarity, life scientists who hold to the evolutionary paradigm assume that ERVs have been introduced into genomes through a process (endogenization) without actually observing the process. But, given the emerging evidence that ERVs protect genomes from retroviral infections—thanks to the koala studies—life scientists who hold to a creation model/intelligent design perspective could reasonably view some of the ERV sequences as features of the genome that were intentionally designed.
Resources
- Who Was Adam? A Creation Model Approach to the Origin of Humanity, 2nd exp. ed. by Fazale Rana with Hugh Ross (book)
- “Questioning Evolutionary Presuppositions about Endogenous Retroviruses” by A. J. Roberts (article)
- “A Common Design View of ERVs Encourages Scientific Investigation” by A.J. Roberts (article)
- “Archetype or Ancestor? Sir Richard Owen and the Case for Design” by Fazale Rana (article)
Endnotes
- Ulrike Löber et al., “Degradation and Remobilization of Endogenous Retroviruses by Recombination during the Earliest Stages of a Germ-Line Invasion,” Proceedings of the National Academy of Sciences USA 115 (August 21, 2018): 8609–14, doi:10.1073/pnas.1807598115.
- Clare Lynch and Michael Tristem, “A Co-Opted Gypsy-Type LTR-Retrotransposon Is Conserved in the Genomes of Humans, Sheep, Mice, and Rats,” Current Biology 13 (September 2, 2003): 1518–23, doi:10.1016/S0960-9822(03)00618-3.






