Long Noncoding RNAs Extend the Case for Creation
I don’t like to think of myself as technology-challenged, but I am beginning to wonder if I just might be. As a case in point, I have no clue about all the things my iPhone can do. It isn’t uncommon for someone (usually much younger than me) to point out features of my iPhone that I didn’t even know existed. (And, of course, there is the TV remote—but that will have to serve as material for another lead.)
The human genome is a lot like my iPhone. The more the scientific community learns about it, the more complex it becomes and the more functionality it displays—functionality about which no one in the scientific community had a clue. It has become commonplace for scientists to discover that features of the human genome—long thought to be useless vestiges of an evolutionary history—actually serve a critical role in the structure and function of the genome.
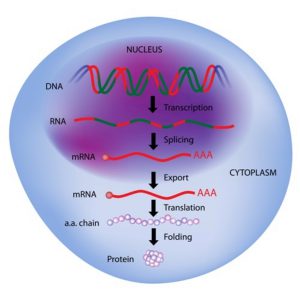
Long noncoding RNAs (lncRNAs) illustrate this point nicely. This broad category of RNA molecules consists of transcripts (where genetic information is transferred from DNA to messenger RNA) that are over 200 nucleotides in length but are not translated into proteins.
Though numbers vary from source to source, estimates indicate that somewhere between 60 to 90 percent of the human genome is transcribed. Yet only 2 percent of the genome consists of transcripts that are directly used to produce proteins. Of the transcripts that are untranslated, researchers estimate that somewhere between 60,000 to 120,000 of the transcripts are noncoding RNAs. Researchers categorize these transcripts as microRNAs (miRNAs), piwi-interacting RNAs (piwiRNAs), small interfering RNAs (siRNAs) and lncRNAs. The first three types of RNAs are relatively small in size and play a role in regulating gene expression.
Figure 1: Transcription and Translation. Image credit: Shutterstock
Initially, researchers thought for the most part that lncRNAs were transcriptional noise—junk. But this view has changed in recent years. Evidence continues to accrue demonstrating that lncRNAs play a wide range of roles in the cell.1 And as evidence for the utility of lncRNAs mounts, the case for the design of the human genome expands.
The Functional Utility of Long Noncoding RNAs
As it turns out, lncRNAs are extremely versatile molecules that can interact with: (1) other RNA molecules, (2) DNA, (3) proteins, and (4) cell membranes. This versatility opens up the possibility that these molecules play a diverse role in cellular metabolism.
Recently, Harry Krause, a molecular geneticist from the University of Toronto, published two review articles summarizing the latest insights into lncRNA function. These insights, including the four to follow, demonstrate the functional pervasiveness of the transcripts.
lncRNAs regulate gene expression. lncRNAs influence gene expression by a variety of mechanisms. One is through interactions with other transcripts forming RNA-RNA duplexes that typically interfere with translation of protein-coding messenger RNAs.
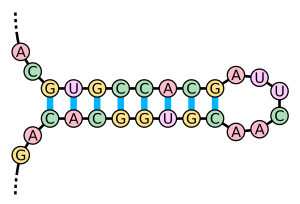
Researchers have recently learned that lncRNAs can also influence gene expression by interacting with DNA. These interactions result in either: (1) a triple helix, made up of two DNA strands intertwined with one RNA strand, or (2) a double helix with the lncRNA intertwined with one of the DNA strands, leaving the other exposed as a single strand. When these duplexes form, the lncRNA forms a hairpin loop that can either indiscriminately or selectively attract transcription factors.
Figure 2: A Hairpin Loop. Image credit: Wikipedia
Though researchers are still learning about the role lncRNAs play in gene regulation, these varied interactions with DNA and proteins suggest that lncRNAs may influence gene expression through a variety of mechanisms.
lncRNAs form microbodies within the nucleus and cytoplasm. A second function recognizes that lncRNAs interact with proteins to form hydrogel-like structures in the nucleus and cytoplasm. These structures are dense and heavily cross-linked subcellular structures that serve as functionally specific regions without a surrounding membrane. (In a sense, the microbodies could be viewed as somewhat analogous to ribosomes, the protein-RNA complexes that synthesize proteins.) In the nucleus, microbodies play a role in transcriptional processing, storage, and stress response. In the cytoplasm, microbodies play a role in storage, processing, and trafficking.
lncRNAs interact with cell membranes. A third role stems from laboratory studies where lncRNAs have been shown to interact with model cell membranes. Such interactions suggest that lncRNAs may play a role in mediating biochemical processes that take place at cell membranes. Toward this end, researchers have recently observed certain lncRNA species interacting with phosphatidylinositol 3,4,5-triphosphate. This cell membrane component plays a central role in signal transduction inside cells.
lncRNAs are associated with exosomes. Finally, lncRNAs have been found inside membrane-bound vesicles that are secreted by cells (called exosomes). These vesicles mediate cell-cell communication.
In short, the eyes of the scientific community have been opened. And they now see the functional importance and functional diversity of lncRNAs. Given the trend line, it seems reasonable to think that the functional range of lncRNAs will only expand as researchers continue to study the human genome (and genomes of other organisms).
The growing recognition of the functional versatility of lncRNAs aligns with studies demonstrating that other regions of the genome—long thought to be nonfunctional—do, in fact, play key roles in gene expression and other facets of cellular metabolism. Most significantly, toward this end, the functional versatility of lncRNAs supports the conclusions of the ENCODE Project—conclusions that have been challenged by some people in the scientific community.
The ENCODE Project
A program carried out by a consortium of scientists with the goal of identifying the functional DNA sequence elements in the human genome, the ENCODE Project, reported phase II results in the fall of 2012. (Currently, ENCODE is in phase IV.) To the surprise of many, the ENCODE Project reported that around 80 percent of the human genome displays biochemical activity—hence, function—with the expectation that this percentage should increase as results from phases III and IV of the project are reported.
The ENCODE results have generated quite a bit of controversy. One of the most prominent complaints about the ENCODE conclusions relates to the way the consortium determined biochemical function. Critics argue that ENCODE scientists conflated biochemical activity with function. As a case in point, the critics argue that most of the transcripts produced by the human genome (which include lncRNAs) must be biochemical noise. This challenge flows out of predictions of the evolutionary paradigm. Yet, it is clear that the transcripts produced by the human genome are functional, as numerous studies on the functional significance of lncRNAs attest. In other words, the biochemical activity detected by ENCODE equates to biochemical function—at least with respect to transcription.
A New View of Genomes
These types of insights are radically changing scientists’ view of the human genome. Rather than a wasteland of junk DNA sequences stemming from the vestiges of an evolutionary history, genomes appear to be incredibly complex, sophisticated biochemical systems, with most of the genome serving useful and necessary functions.
We have come a long way from the early days of the human genome project. When completed in 2003, many scientists at that time estimated that around 95 percent of the human genome consists of junk DNA. That acknowledgment seemingly provided compelling evidence that humans must be the product of an evolutionary history.
Nearly 15 years later the evidence suggests that the more we learn about the structure and function of genomes, the more elegant and sophisticated they appear to be. It is quite possible that most of the human genome is functional.
For creationists and intelligent design proponents, this changing view of the human genome—similar to discovering exciting new features of an iPhone—provides reasons to think that it is the handiwork of our Creator. A skeptic might ask, Why would a Creator make genomes littered with so much junk? But if a vast proportion of genomes consists of functional sequences, this challenge no longer carries weight and it becomes more and more reasonable to interpret genomes from within a creation model/intelligent design framework.
Resources
- “ENCODE Junks Evolutionary Concept” by Fazale Rana (article)
- “Responding to ENCODE Skeptics” by Fazale Rana (article)
- “Do ENCODE Skeptics Protest Too Much? Part 1” by Fazale Rana (article)
- “Do ENCODE Skeptics Protest Too Much? Part 2” by Fazale Rana (article)
- “Do ENCODE Skeptics Protest Too Much? Part 3” by Fazale Rana (article)
- “Do Scientists Accept the Results of the ENCODE Project?” by Fazale Rana (article)
- “Is 75 Percent of the Human Genome Junk DNA?” by Fazale Rana (article)
- “The Human Genome: Copied by Design” by Fazale Rana (article)
- “Protein Binding Sites ENCODEd into the Design of the Human Genome” by Fazale Rana (article)
- Who Was Adam? A Creation Model Approach to the Origin of Humanity, 2nd expanded ed., by Fazale Rana with Hugh Ross (book)
Endnotes
- Allison Jandura and Henry M. Krause, “The New RNA World: Growing Evidence for Long Noncoding RNA Functionality,” Trends in Genetics 33 (October 1, 2017): 665– 76, doi:10.1016/j.tig.2017.08.002; Henry M. Krause, “New and Prospective Roles for lncRNAs in Organelle Formation and Function,” Trends in Genetics 34 (October 1, 2018): 736–45, doi:10.1016/j.tig.2018.06.005.