Becoming Human—Fast!
by Patricia Babin
While recording a recent episode of the I Didn’t Know That! podcast, my colleague Fuz Rana responded to a listener who wanted to know if there is enough time (from an evolutionary perspective) between the divergence of chimps and humans from their common ancestors for humans to have evolved. In other words: Is the rate of evolution sufficient to explain the differences between humans and chimps?
I’ll paraphrase Fuz’s response. By definition, there’s enough time because the evolutionary rate is calculated by determining the number of changes between the common ancestor of humans and chimps and then dividing that number by 6 million years—the amount of time that has elapsed since the divergence of humans and chimps. To put it another way, “the deck is rigged.” So, of course, there’s enough time (from the evolutionary perspective) for humans to have evolved from chimps. Or is there? Perhaps not always, even when using a rigged deck.
Recently, scientists have discovered examples of “accelerated evolution,” that is, a rate of change in the DNA of a species that far exceeds the expected rate. The number of articles using the term “accelerated evolution” was very low in the 1990s, averaging three per year. Between 2000 and 2005, the number increased to an average of 12 per year. Since 2005, the average has doubled to 27 per year. Clearly, the availability of more DNA sequence data is contributing to this trend.
My favorite example of so-called “accelerated evolution” is a section of human RNA dubbed HAR1, for “human accelerated region 1.”1 HAR1 was discovered by a multinational team of scientists. The team focused their efforts on noncoding DNA in hopes of finding the regions responsible for what makes humanity unique from other species. They began by searching the chimpanzee, mouse, and rat genomes for areas of noncoding DNA 100 base pairs long and at least 96 percent conserved (unchanged) between the three species. (Sequence conservation is to be expected for DNA regions with important functions). Their search revealed 35,000 such sequences in the three species.
The team then compared these DNA regions to the equivalent sequence in an additional seven species in order to identify stretches of DNA conserved across a wider range of species—including animals as diverse from chimps as chickens and the platypus. Finally, they compared these highly conserved sequences to the same regions in humans to identify those that had changed in humans and, therefore, might be implicated in helping develop unique human characteristics. This process resulted in identifying 49 noncoding regions of human DNA that are highly conserved among other amniotes but differ significantly in humans. These regions (labeled HARs) are good candidates for explaining some of humanity’s biological uniqueness.
Image source: Bobisbob
The HAR1 region showed the greatest variance in humans. It is a 118 base pair DNA segment with 18 bases that differ from the same region in the other species. From an evolutionary point of view less than one (0.27) base change(s) is expected in the HAR1 DNA sequence in the time elapsed since chimps and humans diverged from their common ancestor. Eighteen changes in human HAR1 equates to 67 times the number of changes expected.
Additionally, the equivalent of HAR1 in all those other species is very highly conserved. The researchers report that “only two bases (out of 118) are changed between chimpanzee and chicken.” There are some attempts in the scientific literature to explain the phenomena of “accelerated evolution.” However, HAR1 doesn’t fit into the explanations being offered2 because it is so highly conserved.3 In addition to the astonishing number of changes in human HAR1, the changes reflect GC-biased conversion, another unexpected phenomena in which adenine (A) or thymine (T) change consistently to cytosine (C) or guanine (G).
The HAR1 DNA region produces HAR1 RNA. Scientists have demonstrated that HAR1 RNA is expressed selectively in Cajal-Retzius neurons (cells that assist in brain development) in the fetal human cerebral cortex from 7 to 19 weeks of gestation.3 After that time, HAR1 RNA is expressed in other cells of the developing human brain. This kind of selective expression with regard to both location and time during development generally indicates functionality. So it is likely that the RNA is playing an important role in brain development.
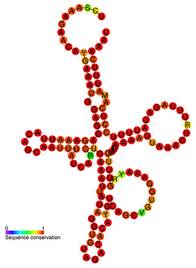
One of the ways in which RNA differs from DNA is that while DNA typically has only a few standard shapes, RNA molecules can fold into a wide diversity of enormously complex shapes critical to functionality (for example, see figure 2). Since HAR1 RNA is likely an important molecule, it is not surprising that scientists are publishing competing models for the structure of this particular RNA.4
Figure 2. Secondary structure of human HAR1 RNA
Image source: User:Ppgardne
So, if you are known for really bad jokes, you could playfully pose the following question to your evolutionist friends, “What do a chimp, a platypus, and a human have in common?” The answer to the riddle is “A lot, but not HAR1.” Hey, it’s a non-threatening way to start a discussion.
Endnotes
- Katherine S. Pollard et al., “An RNA Gene Expressed during Cortical Development Evolved Rapidly in Humans,” Nature 443 (September 14, 2006): 167–72.
- David S. Lawrie, Dmitri A. Petrov, and Philipp W. Messer, “Faster than Neutral Evolution of Constrained Sequences: the Complex Interplay of Mutational Biases and Weak Selection,” Genome Biology and Evolution 3 (January 2011): 383–95.
- Katherine S. Pollard et al., “An RNA Gene Expressed,” 167–72.
- Katherine S. Pollard et al., “An RNA Gene Expressed,” 167–72; Artemy Beniaminov, Eric Westhof, and Alain Krol, “Distinctive Structures between Chimpanzee and Human in a Brain noncoding RNA,” RNA 14 (July 2008): 1270–75.