“Junk” DNA: An Outdated Concept, Part 2 (of 6)
by Dr. Patricia Babin
In the first article in this series, I recalled my experience of hearing a biologist use the presence of Alu elements (a type of so-called “junk DNA”) in the human genome as a “proof” of evolution. My natural skepticism prompted me to look into the biologist’s claims. Even a preliminary review of the research revealed that supposed “junk DNA” isn’t such a wise choices for proving evolution.
More recently, as an RTB visiting scholar, I found myself with an opportunity to delve farther into Alu elements research as I prepared for a podcast on the topic. The depth and breadth of the literature ascribing functionality to Alu elements astonished me. (A search of the National Library of Medicine for articles on the functions of Alu elements identifies over 1,600 articles in peer-reviewed scientific journals. The same search performed on Google Scholar1 reflects what I found in the National Library of Medicine.)
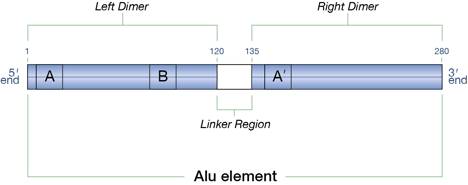
Like the functions of any portion of DNA, the functions of Alu elements are based on its sequence. Some sequences are found so often in DNA and are so foundational to their function that scientist have labeled them “motifs.” A typical Alu element is approximately 300 base pairs long and contains a number of these motifs (figure 1):
Figure 1: An Alu element
- Inverted repeat structure: The left dimer of the Alu element is complementary to the right dimer, which allows one dimer to base pair with another to form a helix. This type of structure is called an inverted repeat structure. Though the base pairing between the two portions does not occur in DNA, it can form in the RNA made from the DNA.
- Box A, Box B, and Box A’: Box A and Box B are control sequences. A specific protein binds to them, initiating transcription by RNA polymerase III. Box A’ is the complement of Box A.
- Nucleosome binding sequences: Numerous studies have demonstrated that Alu elements possess the ability to stimulate nucleosome formation within the Alu element.2 Up to two nucleosomes can be accommodated per Alu element.
- Transcription factor binding sites: Numerous transcription factors, perhaps as many as 66, are thought to bind to Alu elements.3
- Splice signals: Some Alu elements experience minor changes in their sequences that can result in the inclusion of the Alu element into messenger RNA (mRNA) during the splicing process. Researchers understand how an Alu element changes so that it contains (splice sites), the sequences that mark the beginning and end of a segment included in mRNA . Yet, they lack a firm understanding of the other sequences within a DNA segment that creates the broader context in which splicing occurs. However, whatever those sequences are, they occur in some Alu elements.4
- Missing “stop” signal for RNA Polymerase III: This motif’s absence is of significant importance. The presence of Box A and Box B and the absence of a strong “stop” signal for RNA Polymerase III means that the DNA located adjacent to the end of the Alu element will frequently be copied into RNA.5
- Conserved sequence: GAGGCTGAGG is a conserved sequence whose function is not yet known. Though scientists don’t understand the function of this sequence, the fact that it is so highly conserved indicates that it has an important function.
In addition to these motifs found within Alu elements, the elements’ location in the genome is critical for their function. Though there are over a million copies of Alu elements scattered throughout the human genome, their locations are not random. Alu elements are:
- concentrated in gene rich regions of the chromosome, particularly near metabolic genes;6
- concentrated 4,500 base pairs before the transcription start site of genes;.7
- often located upstream of important DNA sequences that could not be transcribed were it not for certain motifs found in Alu elements;8 and
- often located near genes regulated by the p53 tumor suppressor gene, one of the most important regulators of gene expression.9
Figure 2. Human chromosomes stained to show Alu elements (lime green color). Image credit: PLoS Biology.
The next installment of this series will focus on the functions of Alu elements in detail.
| Part 1 | Part 2 | Part 3 | Part 4 | Part 5 | Part 6 |
Endnotes
- The preceding link presents the results of a Google Scholar search for the terms “Alu element” and “function.” (These results may contain duplicates and non-scholarly works.) Of the 18,800 entries, 14,000 of them were published between 2000 and 2011. Searching for the terms “Alu element” and “function” in the National Library of Medicine identifies 1,622 articles published in peer-reviewed scientific journals.
- Yoshiaki Tanaka et al., “Effects of Alu Elements on Global Nucleosome Positioning in the Human Genome,” BMC Genomics 11 (May 17, 2010): 309–19; Sheila M. Reynolds, J. A. Bilmes, and W. S. Noble,, “Learning a Weighted Sequence Model of the Nucleosome Core and Linker Yields More Accurate Predictions in Saccharomyces cerevisiae and Homo sapiens,” PLoS Computational Biology 6 (January 8, 2010): 1–17.
- Paz Polak and Eytan Domany, “Alu Elements Contain Many Binding Sites for Transcription Factors and May Play a Role in Regulation of Developmental Processes,” BMC Genomics 7 (June 1, 2006): 133–47.
- Shihao Shen et al., “Widespread Establishment and Regulatory Impact of Alu Exons in Human Genes,” Proceedings of the National Academy of Sciences, USA 108 (February 15, 2011): 2837–42.
- Glen Borchert, W. Lanier, and B. L. Davidson, “RNA Polymerase III Transcribes Human microRNAs,” Nature Structural & Molecular Biology 13 (December 2006): 1097–1101.
- Polak and Domany, “Alu Elements,” 133–47.
- Ibid.
- Borchert, Lanier, and Davidson, “RNA Polymerse III,” 1097–1101.
- Feng Cui, M. V. Sirotin, and V. B. Zhurkin, “Impact of Alu Repeats on the Evolution of Human p53 Binding Sites,” Biology Direct 6 (2011): 2–20.