“Junk” DNA: An Outdated Concept, Part 5 (of 6)
by Patricia Babin
For the last four weeks, this article series has detailed the junk DNA argument against intelligent design and shown why this argument no longer holds water. Last week, I focused exclusively on transcription factor binding to Alu elements (a type of so-called “junk” DNA once thought to be functionless) because of the critical functions and diversity of transcription factors involved.
In this post, I’ll highlight four additional functions of Alu elements through which gene expression can be regulated. The first function is a specialized form of transcription; the others occur after transcription is completed but before translation begins.
1. Generating miRNA
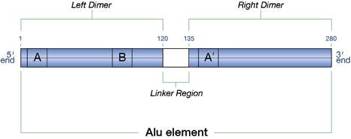
RNA Polymerase II (RNAP II) transcribes the coding regions of DNA (segments containing the information necessary to create proteins) to create mRNA. Alu elements contain control sequences, known as Box A and Box B (see figure 1) for a different RNA polymerase—RNA Polymerase III (RNAP III). RNAP III transcribes portions of the DNA—dubbed housekeeping genes—that generate functional RNAs used abundantly in the cell; which means they must be transcribed more frequently than an average gene. Housekeeping genes are under less strict control than genes transcribed by RNAP II presumably because of the need to transcribe them so frequently.
Figure 1
Yet although Alu elements contain control regions for RNAP III, they lack an important structural motif associated with this molecule—a stop signal. This means that once RNAP III begins transcribing the Alu element, it continues on past the end of the Alu element DNA, making an RNA version of whatever is located beyond it. Some of the RNA transcripts generated by this method undergo several processing steps to eventually produce a short RNA—about 22 nucleotides in length—known as miRNA (microRNA).
A recent study1 identified 24 miRNAs transcribed because of their proximity to an Alu element. Since expression of miRNA is both tissue- and developmental-stage specific, the scientists who conducted this study expect to find more miRNAs as they examine more tissues. Furthermore, miRNAs have been shown to regulate transcription and translation through a variety of means and may be the most abundant regulatory factor in the human genome.2
2. A Target for mRNA Editing
In part 2 of this series, I discussed inverted repeat structure, a structural motif in Alu elements. This motif allows mRNA-containing Alu elements to fold into a double helix structure. The presence of the double helix causes the mRNA to become a target for several important enzymes. (The double helix can be formed by the Alu element folding back on itself or through base pairing with another mRNA that contains an Alu element.) One of these is ADAR (“adenosine deaminase acting on RNA”). This enzyme converts adenosine nucleotides in RNA to inosine nucleotides in a process called mRNA editing.
RNA is typically composed of adenosine, guanosine, cytidine, and uridine. When the ribosome encounters inosine, it is treated as guanosine. Therefore, an adenosine-to-inosine transition can create a different codon in mRNA and result in production of a different protein. Several recent studies have demonstrated that Alu elements play a critical role in driving the editing of mRNA transcripts in humans.3 The DNA for the genes that generate these mRNA transcripts is generally identical in humans and primates, but the protein produced by the genes differs in humans and primates because of mRNA editing. Many of these proteins are critical for brain development and function.
3. A Target for Destruction
The presence of a double helix in mRNA containing an Alu element makes the mRNA a target for another protein called STAU1.4 By binding to the mRNA, STAU1 causes the mRNA to be degraded and, therefore, unusable for transcription, meaning no protein will result from the mRNA. This is a newly discovered strategy for regulating gene expression.
4. New Exons
The sequences within mRNA that direct the process of mRNA splicing are understood to a limited extent. In addition to sequences within the mRNA, other molecules in specific tissue types are probably also involved in directing how splicing occurs. Though many aspects of splicing are not well understood, it is clear that Alu elements contain sequences that cause alternative splicing to occur. Thousands of human mRNA transcripts have been shown to contain sections derived from Alu elements during the process of splicing.5 These new sections are called exons.
The presence of these new exons in the mRNA results in a different protein being produced in humans (when compared to other primates). Scientists have recently discovered that mRNAs that code for transcription factors containing a special motif called zinc fingers frequently contain exons derived from Alu elements. Since zinc finger transcription factors are major regulators of transcription, Alu element involvement here has a major impact on gene expression. Additionally, mRNAs that contain exons derived from Alu elements have also been demonstrated to produce less protein as a result.
These four functions of Alu element gene expression––all recent discoveries––add to the weight of evidence demonstrating function for “junk” DNA. This concept seems as outdated as a typewriter.
Next week I’ll close out this series with some concluding thoughts on Alu elements.
| Part 1 | Part 2 | Part 3 | Part 4 | Part 5 | Part 6 |
Endnotes
- Tong J. Gu et al., “Alu Directed Transcriptional Regulation of some Novel miRNAs,” BMC Genomics 10 (2009): 563–574.
- John S. Tsang, Margaret S. Ebert, and Alexander van Oudenaarden, “Genome-wide Dissection of MicroRNA Functions and Co-Targeting Networks Using Gene Set Signatures,” Molecular Cell 38 (2010): 140–53.
- Nurit Paz-Yaacov et al., “Adenosine-to-Inosine RNA Editing Shapes Transcriptome Diversity in Primates,” Proceedings of the National Academy of Sciences 107 (June 21, 2010): 12174–179; Sivan Osenberg et al., “Alu Sequences in Undifferentiated Human Embryonic Stem Cells Display High Levels of A-to-I RNA Editing,” PLoS One 5 (2010): 1–11.
- Chenguang Gong and Lynne E. Maquat, “lncRNAs Transactivate STAU1-Mediated mRNA Decay by Duplexing with 3′ UTRs via Alu Elements,” Nature 470 (February 10, 2011): 284–88.
- Shihao Shen et al., “Widespread Establishment and Regulatory Impact of Alu Exons in Human Genes,” Proceedings of the National Academy of Sciences 108 (February 15, 2011): 2837–42.